The MetaDome web server
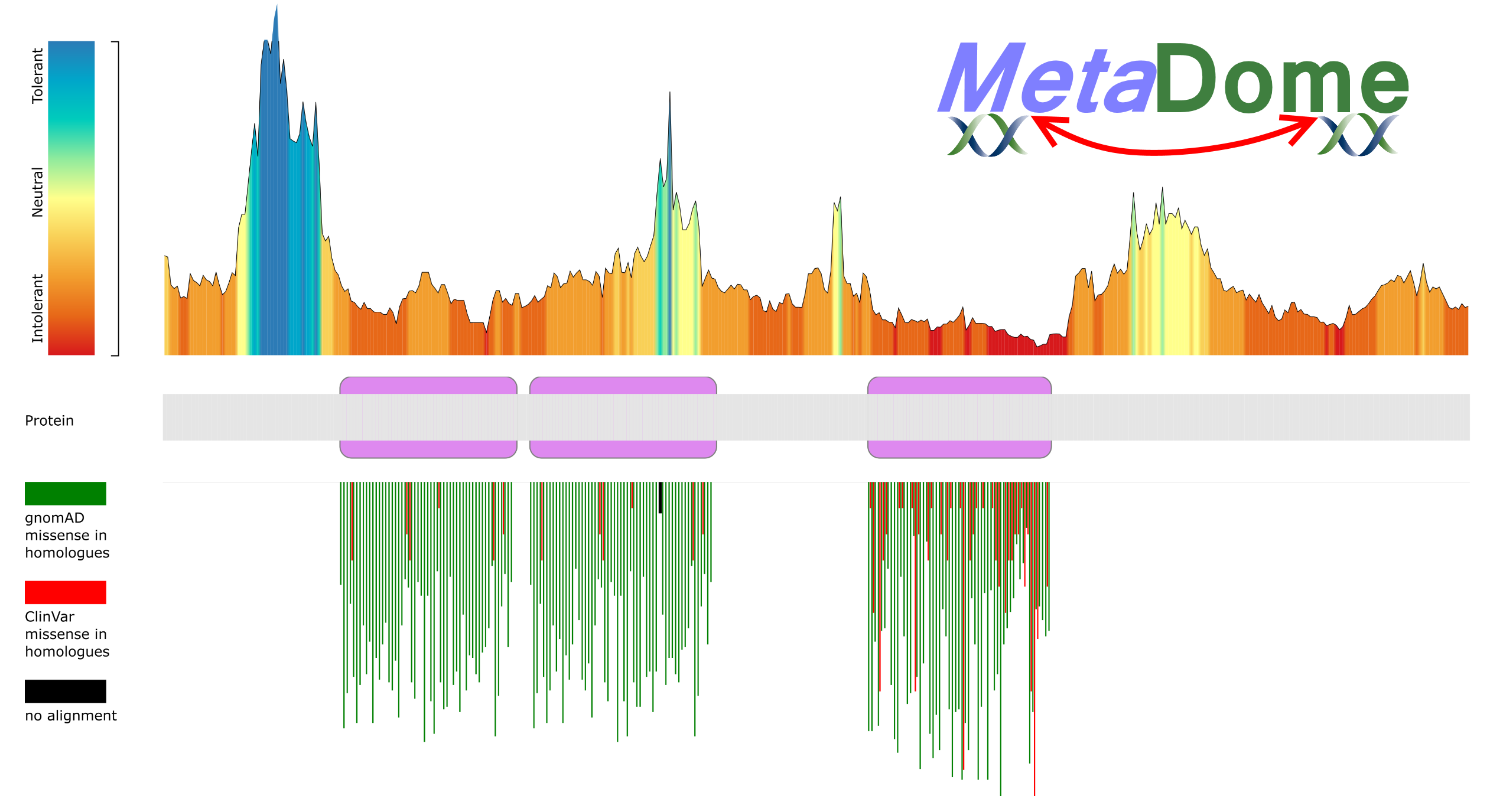 MetaDome is aimed at professionals in the (bio-)medical field of human genetics who wish to visualize the position of their mutation of interest in the context of general population-based genetic variation and provide detailed information of pathogenic variants found across homologous domain positions.
MetaDome is aimed at professionals in the (bio-)medical field of human genetics who wish to visualize the position of their mutation of interest in the context of general population-based genetic variation and provide detailed information of pathogenic variants found across homologous domain positions.
How to access
- The MetaDome web server is freely accessible
- The code is open-source available on GitHub
- Raw data available at Zenodo.
Citing MetaDome
- L. Wiel, et al. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Human Mutation, May 2019.
Latest Publications
- L. Wiel*, J.E Hampstead*, et al. de novo mutation hotspots in homologous protein domains identify function-altering mutations in neurodevelopmental disorders. The American Journal of Human Genetics, January 2023
- J. Kaplanis*, K.E. Samocha*, L. Wiel*, Z. Zhang*, et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature, Oct 2020
- L. Wiel, et al. Aggregation of population‐based genetic variation over protein domain homologues and its potential use in genetic diagnostics. Human Mutation, November 2017.
Notable Achievements
- MetaDome was added as a UCSC Genome Browser human constraint scores track in 2021
- MetaDome was added as a track to MobiDetails in 2020
- The MetaDome publication was a Top Accessed Human Mutation Article in 2019 and Top Cited in 2020
- Editor’s Choice Article of Human Mutation
- Featured on the Front Cover of Human Mutation
